DNA – Ligand interactions and photocontrol:
In recent years, we have focused on studying DNA systems in their double-stranded conformation (B-DNA). Specifically, our research has explored light-induced processes such as photodamage, photostability, and ligand-mediated photosensitization.
As a fundamental study, we have characterized the singlet excited-state manifold of the five canonical DNA/RNA nucleobases in the gas phase using time-dependent density functional theory (TD-DFT) and restricted active space second-order perturbation theory (RASPT2). Our findings demonstrate that cost-effective TD-DFT approaches are well-suited for simulating linear and nonlinear spectroscopies in realistic multichromophoric DNA/RNA systems with both biological and nanotechnological relevance.
B-DNA Photostability
We have investigated various photostability mechanisms occurring in the DNA double strand, including excited-state proton transfer between adjacent nucleobases. Notably, the excited-state hydrogen transfer within guanine–cytosine base pairs has been analyzed from a dynamic perspective using advanced molecular modeling and simulation techniques. Our findings reveal a previously unidentified DNA decay pathway, termed four-proton transfer (FPT). This discovery provides new insights into DNA photostability and its fundamental processes.

Figure adapted from DOI: 10.1039/C8SC03252A
B-DNA Photodamage
In the study of DNA photodamage, we have investigated the interaction of two organic dyes with B-DNA, focusing on their binding modes and photophysical properties that facilitate DNA photosensitization through an electron transfer mechanism. To achieve this, we employed a comprehensive multiscale approach, integrating advanced molecular dynamics simulations with hybrid quantum mechanics/molecular mechanics (QM/MM) methods. This methodology can be readily extended to other B-DNA ligands to assess their potential for inducing photodamage.
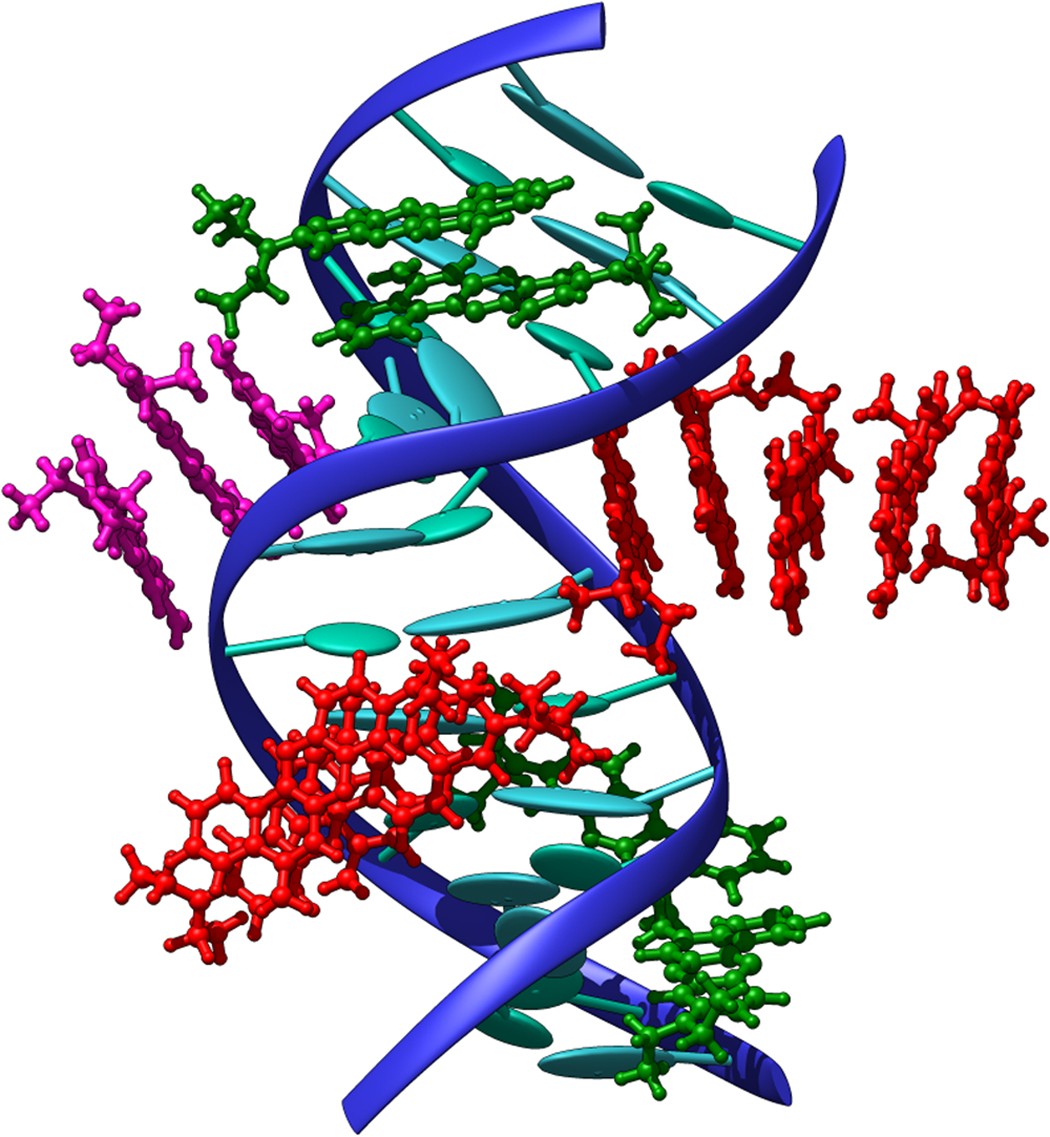
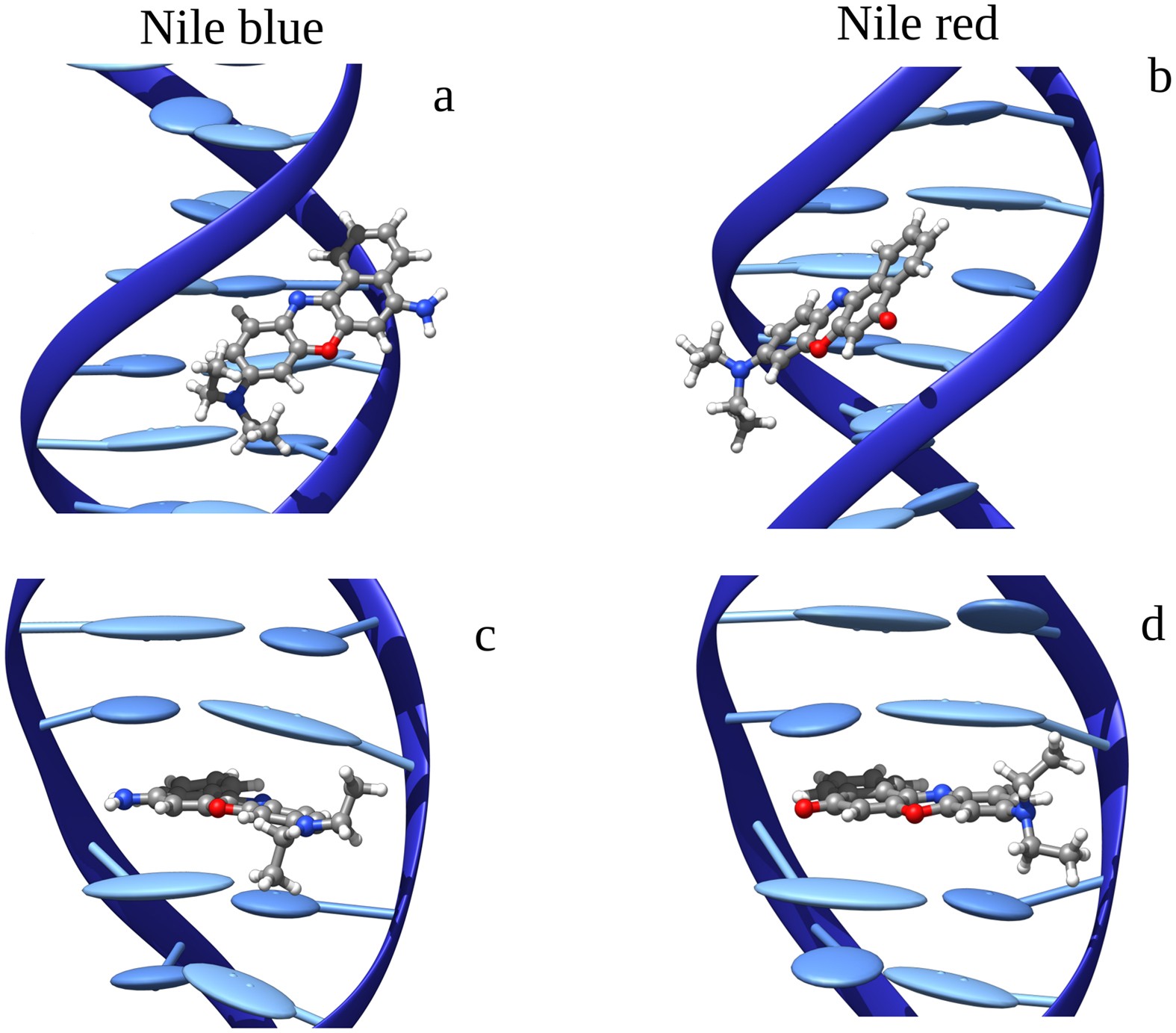
Figures adapted from DOI: 10.1038/srep28480
Additionally, we have conducted computational studies on the hydrogen abstraction process induced by triplet benzophenone on thymine nucleobases and the DNA backbone sugar. To investigate this reaction, we employed a combination of high-level multiconfigurational perturbation theory and density functional theory. Furthermore, the behavior of benzophenone within the DNA environment was explored using molecular dynamics simulations and hybrid quantum mechanics/molecular mechanics (QM/MM) methods.
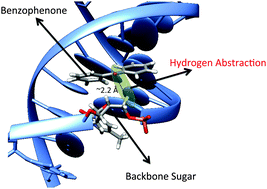
Figure adapted from DOI: 10.1039/C5CP07938A
B-DNA Photosensitization
Furthermore, we have explored the potential therapeutic application of combining photoactive drugs with visible light irradiation through DNA photosensitization. Our approach involves simulating both the linear and non-linear optical properties of the drug, as well as its interaction with DNA, taking into account dynamic and vibrational effects. Specifically, we employed a multiscale molecular modeling and simulation strategy that integrates classical molecular dynamics (MD), quantum chemistry (QM), and hybrid quantum mechanics/molecular mechanics (QM/MM) methods. This comprehensive approach allowed us to investigate the optical properties of BMEMC in complex environments.
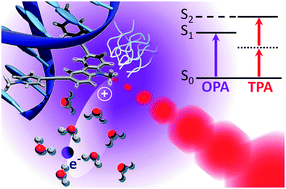
Figure adapted from DOI: 10.1039/C6CP02592G
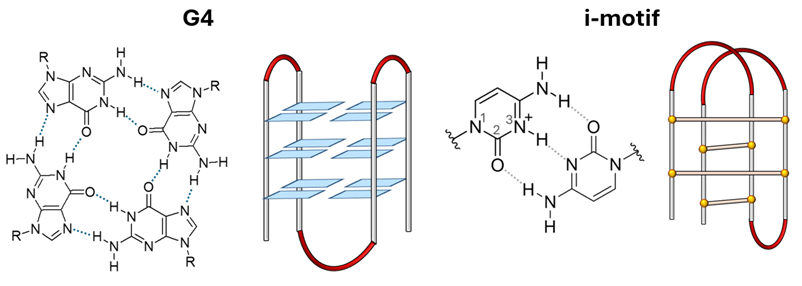
We have recently broadened our research on DNA to encompass non-canonical structures, including G-quadruplexes (G4) and i-motifs. Our specific objectives are:
i) Investigating their structure at the molecular level.
ii) Assessing the stabilizing or destabilizing effects of specific ligands.
iii) Controlling their formation through photochemical activation using photoactive ligands with biologically relevant properties.
To achieve a comprehensive understanding of these systems, an integrated approach combining experimental and computational studies is crucial. Regarding the computational part, docking studies as well as molecular dynamics and hybrid quantum mechanics/molecular mechanics methods are used to obtain a complete picture of the systems and processes under study.